Research
Central themes of our research are development of bioinformatics methods and their application for interpreting genome and transcriptome data. We focus on cancer genomics towards precision medicine and drug target discovery based on integrative analysis of massive public data sets. Deep sequencing, including the single cell techniques, and machine learning are the two prime methods of our lab. Most of these projects are carried out in extensive collaboration with experimental biologists, clinicians, and biotech companies.
Cancer Genome Sequencing
Cancer is a genetic disease caused by accumulation of somatic mutations through lifetime. Recent studies such as the TCGA (The Cancer Genome Atlas) project highlight the heterogeneous nature of cancer, emphasizing the importance of multi-layered omics approach to characterize and dissect the genetic basis of cancer. We produce diverse types of omics data such as whole genome, exome, transcriptome sequencing using cancer patient samples. Those data are analyzed in combination with the massive public data to identify novel genomic aberrations of driver potential and novel patient subtypes. Cancer types we studied so far include lung adenocarcinoma from never-smokers, gastric cancer of early-onset, T cell lymphoma, triple negative breast cancer, and glioma.

Modeling Immunotherapy Responses
Immune checkpoint blockades (ICBs) are revolutionizing the cancer therapy with durable responses in 20-30% of patients. We are developing a computational model to predict the responses to anti-PD-1 treatment. In collaboration with Prof. Se Hoon Lee at the Samsung Medical Center, we analyzed the exome and transcriptome data from >150 lung cancer patients who received the anti-PD-1 treatment. Our machine learning model achieved the highest accuracy with AUC = 0.91. We also analyzed the single cell as well as bulk RNA-seq data from the mouse PDX models that received the anti-PD-1 treatment. Time-series data reveals a number of target cell types relevant to the efficacy of ICBs.


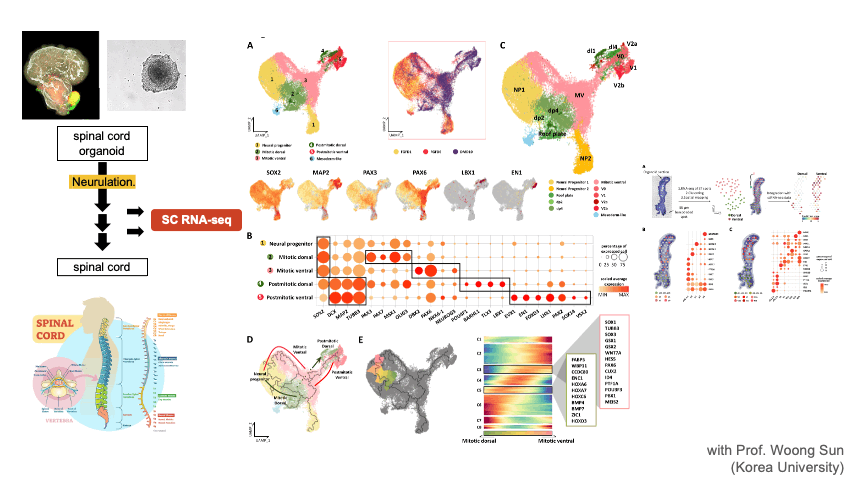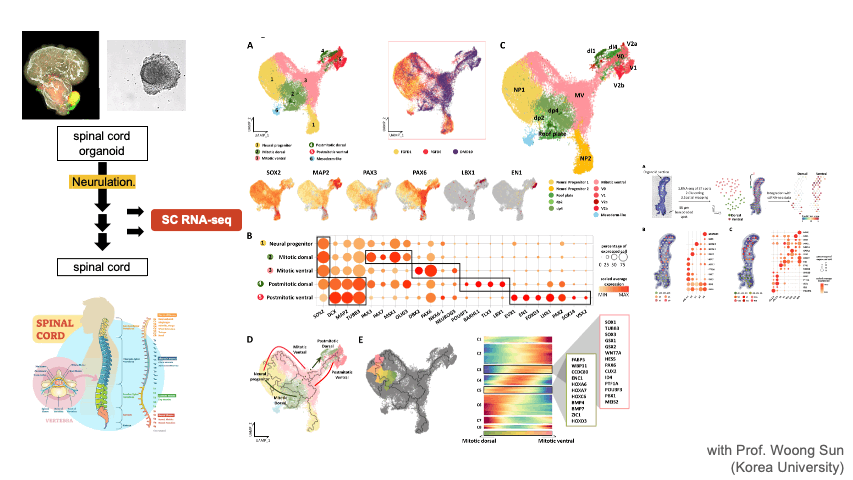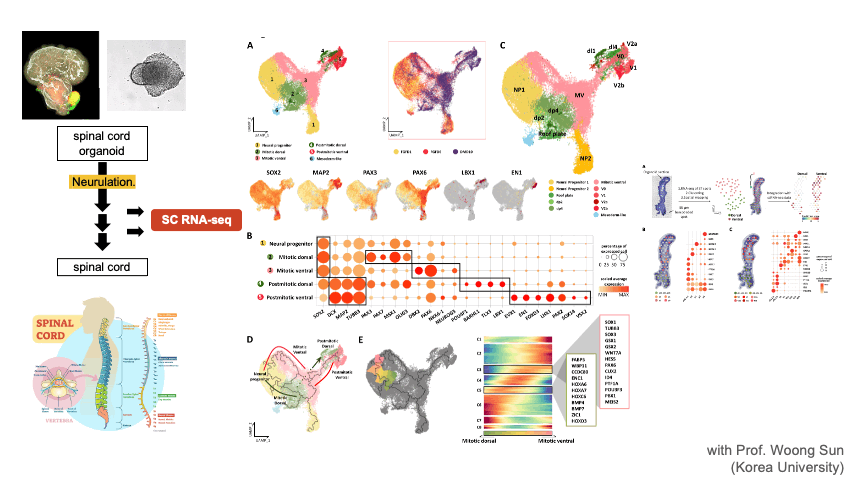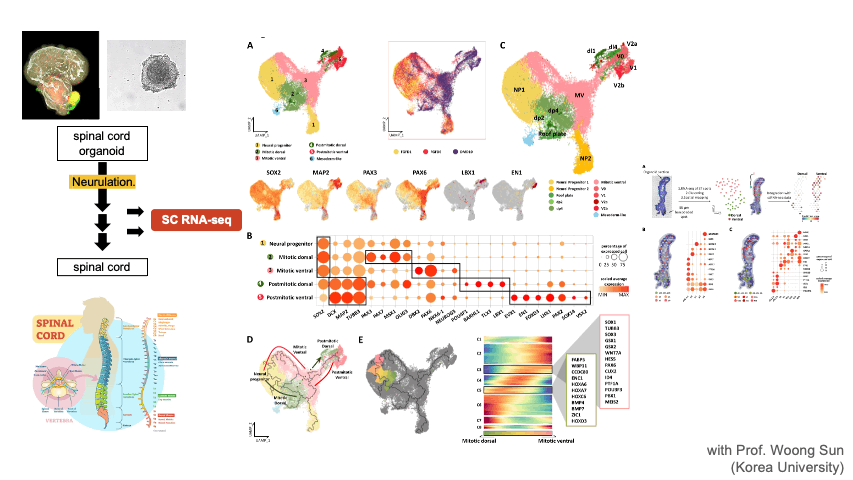
Single Cell RNA-seq & Spatial Transcriptomics
Single cell techniques provide new opportunities to dissect the cell types and composition at an unprecedented resolution. We apply these new techniques to solve diverse biological problems including cancer immunotherapy, tumor metastasis, CNS organoid development, and disease mechanisms. We are also developing a database of single cell CNS organoids by aggregating relevant public data sets.

Drug AI
Chemical perturbation data have proved to be valuable resources of identifying drug targets. Additionally, massive genetic screening is routinely performed using RNAi and CRISPR techniques. LINCS and DepMap are the two representative data sets of chemical and genetic perturbations, respectively. By combining the two perturbation data sets with molecular characteristics, available in the CCLE, we are developing algorithms to identify novel drug targets and combination treatments.

